Welcome to My Page
As a Senior Machine Learning Engineer at Datarock, I pioneer the development of advanced AI solutions tailored for the mining sector. My role involves leveraging ML and AI algorithms to analyze both structured geological data (such as geophysical and geochemical data) and unstructured data (including seismic and geophysical images).
At Datarock, I’ve been at the forefront of spearheading cutting-edge AI methodologies, encompassing self-supervised learning, open-set recognition, graph neural networks, and innovative techniques like inpainting using latent diffusion models and image registration, all finely tuned for geological datasets. My expertise extends to optimizing computational processes and data management tasks through Amazon Web Services (AWS), where I’ve delved deep into AWS intricacies for enhanced efficiency.
You can explore a selection of my projects on GitHub, each accompanied by a concise overview of its purpose and significance.
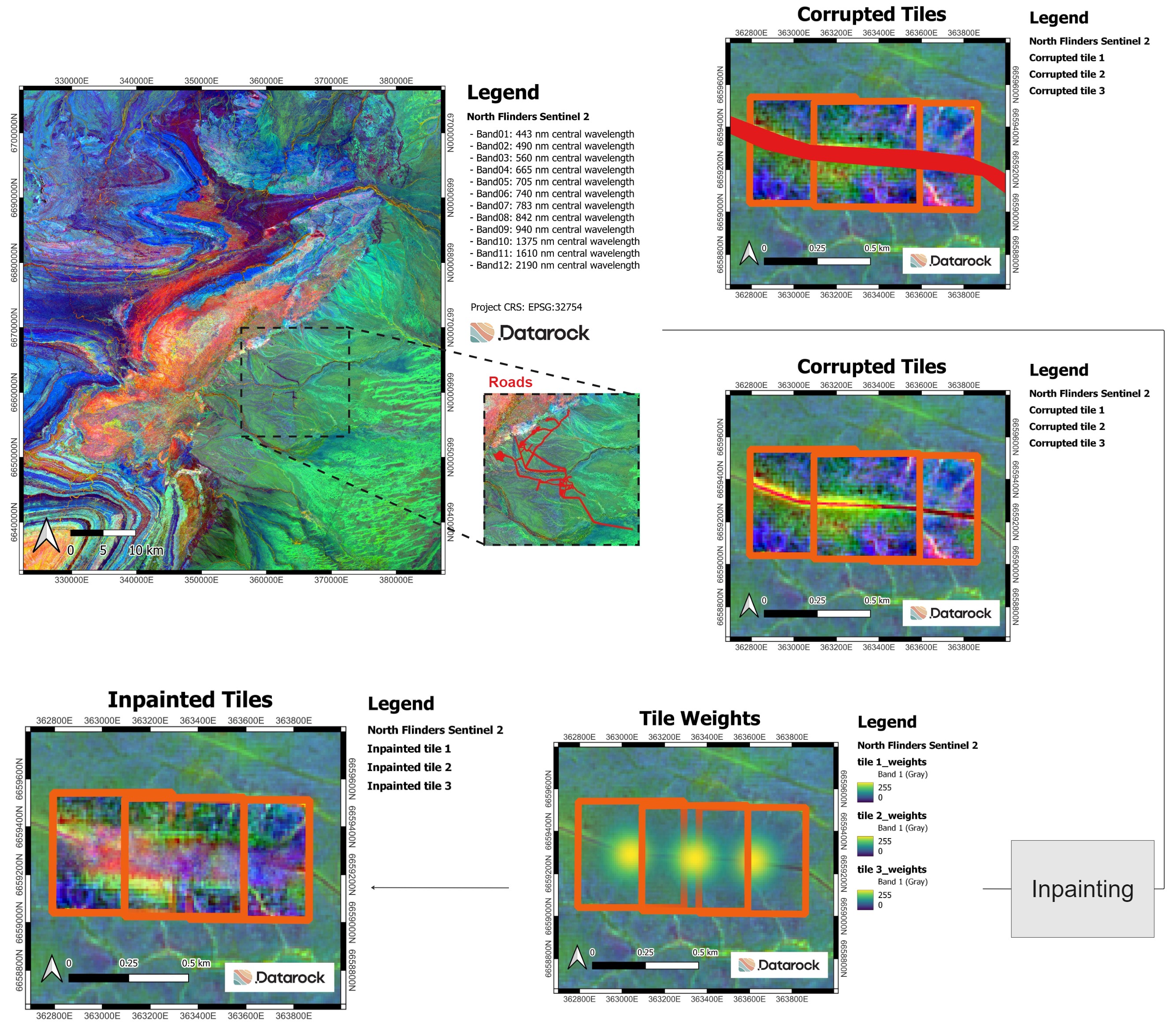
Enhancing Geological Mapping Through Inpainting of Surface Infrastructure Artefacts
This project underscores the significance of inpainting methodologies in enhancing the accuracy and reliability of geological mapping based on aerial and satellite imagery. By effectively removing surface infrastructure artefacts and inpainting missing spatial data, these techniques enable more precise prospectivity mapping and facilitate the advancement of geological understanding and exploration strategies in diverse landscapes.
This work is accepted for oral presentation at the Artificial Intelligence for Geological Modelling and Mapping conference.
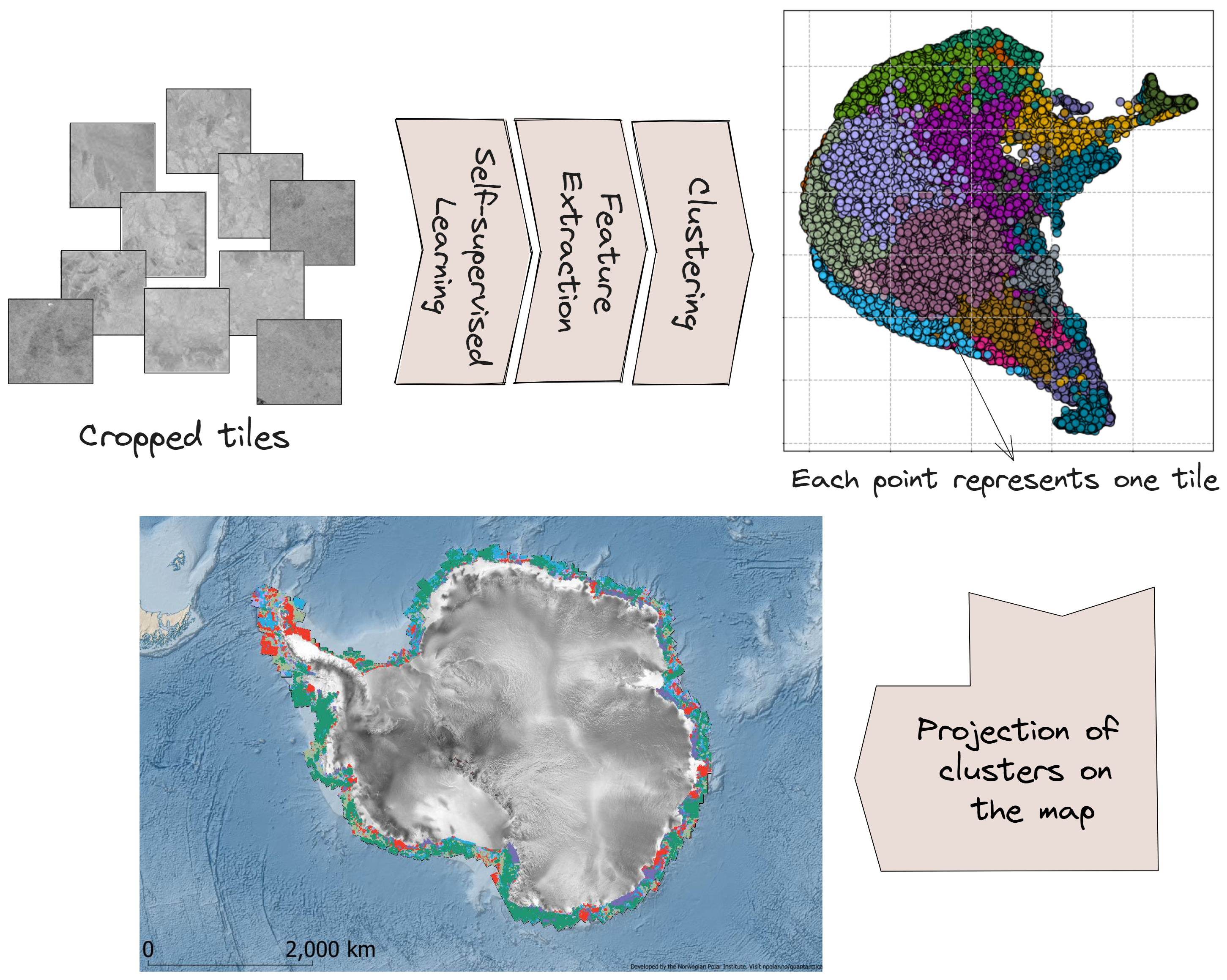
Self-Supervised Learning on the Coasts of Antarctica: A Fun Project
In this fun project, we employed self-supervised learning to train a model on the data of the coasts of Antarctica. Our objective was to identify regions along the Antarctic coasts with similar geological characteristics. The model training focused solely on the coastal areas. Upon training the model on the coasts of Antarctica, features were extracted for each tile and passed to a clustering algorithm. The clustering process assigned each tile to a cluster, with different colors representing distinct clusters. These clusters were then projected back onto the map, revealing regions with similar geological characteristics.
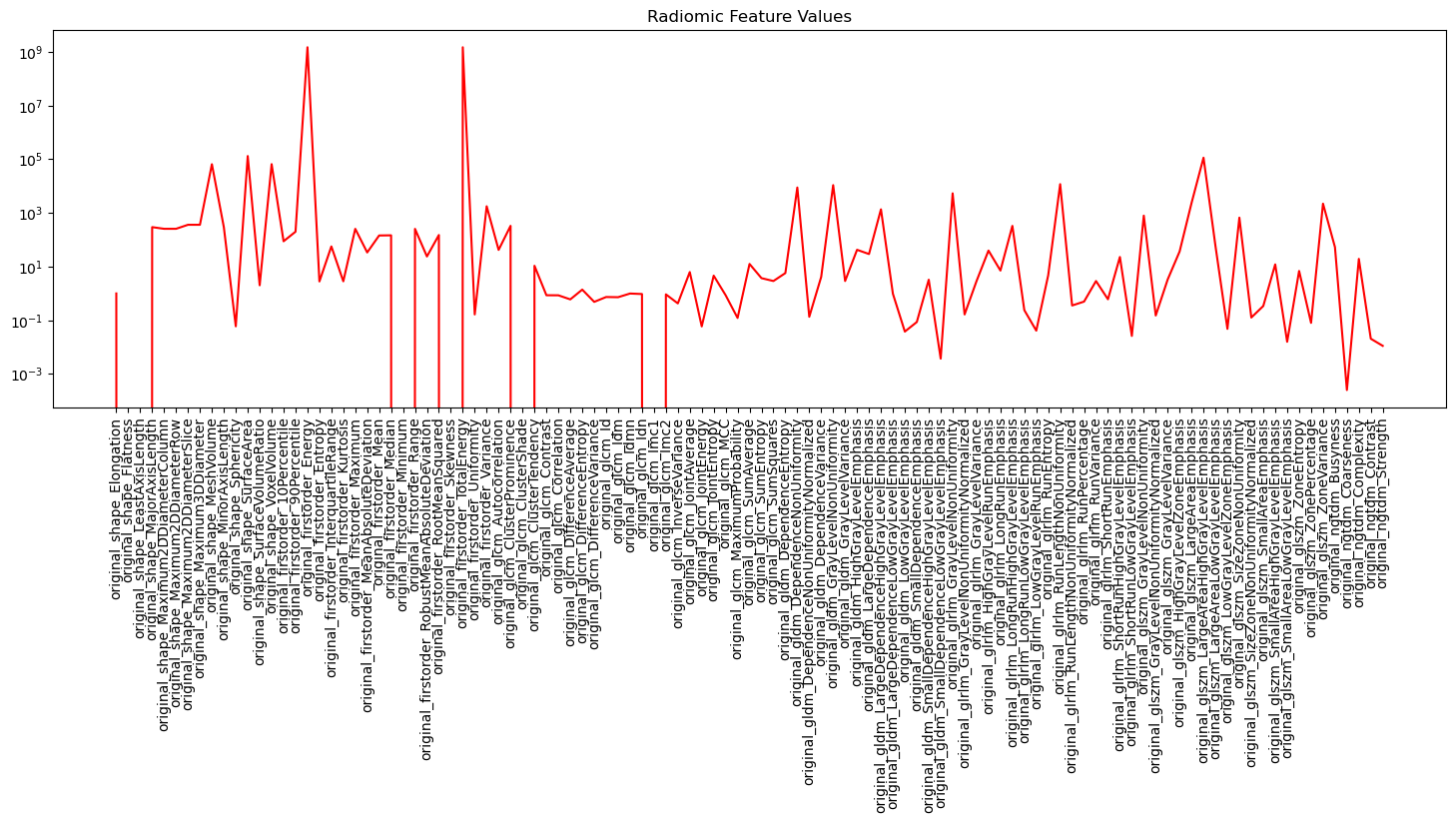
Using Radiomics for Tree Bark Identification
Since the term radiomics was first coined in 2012, it has widely been used for medical image analysis. Radiomics is a method that automatically extracts a large of number of different features from medical images. These features have been able to uncover characteristics that can differentiate tumoral tissue from normal tissue and tissue at different stages of cancer.
In this project, we aim to show that radiomic features can be useful for analysis of images in other domains as well. As a example, we show that radiomic features can be used for tree bark identification. We use a public dataset of tree bark images which can be found here. All the code is provided in the RadiomicsTutorial repository.
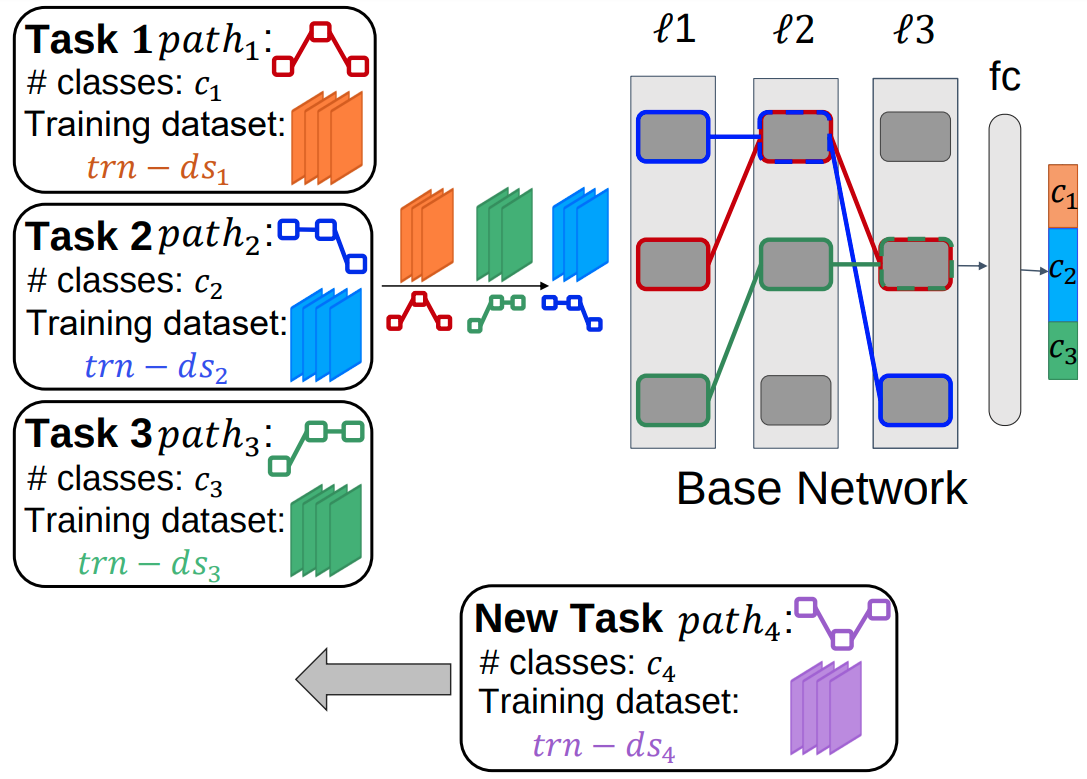
Parallel Learning for Robust and Transparent AI
In this project, we develop an algorithm for training a DL model on multiple tasks in a continual learning scenario. In contrast to the proposed algorithms where the DL model is trained on tasks sequentially, our proposed algorithm trains the DL model on the existing tasks in parallel. The parallel approach allows a DL model to learn generalizable representations and eliminates the problem of catastrophic forgetting which is common in sequential learning approaches. The diagram of the proposed algorithm is shown below.
The code is available at the link below.
https://github.com/MahsaPaknezhad/PaRT-ParallelLearningForRobustAndTransparentAI
Please cite our paper:
Paknezhad, M., Rengarajan, H., Yuan, C., Suresh, S., Gupta, M., Ramasamy, S., & Kuan, L. H. (2022). PaRT: Parallel Learning Towards Robust and Transparent AI. arXiv preprint arXiv:2201.09534.
A Framework for Training Adversarially Robust Models With a Data Sparcity Hypothesis
Deep learning (DL) models have shown to be susceptible to adversarial attacks. To Mitigate adversarial vulnerability of DL models many algorithms have been proposed. However, a clear explanation for why DL models are susceptible to adversarial attacks is still lacking. We hypothesize that the adversarial vulnerability of DL models stems from two factors: 1) data sparsity and 2) the existence of many redundant parameters in the DL models. Owing to these factors, we believe different models can come up with different decision boundaries with comparably high prediction accuracy and that the appearance of the decision boundaries in the space around the class distributions does not affect the prediction accuracy of the model. We hypothesize that the ideal decision boundary is as far as possible from the class distributions. We developed a training framework to observe if DL models can learn such a decision boundary.
We then measure adversarial robustness of the models trained using this training framework against well-known adversarial attacks and find that models trained using their framework, as well as regularization methods and adversarial training support our hypothesis of data sparsity and that models trained with these methods learn to have decision boundaries more like the aforementioned ideal decision boundary. We also show that the unlabeled data generated by noise is almost as effective on adversarial robustness as unlabeled data sourced from existing datasets or generated by synthesis algorithms. A diagram of our training framework is shown below.
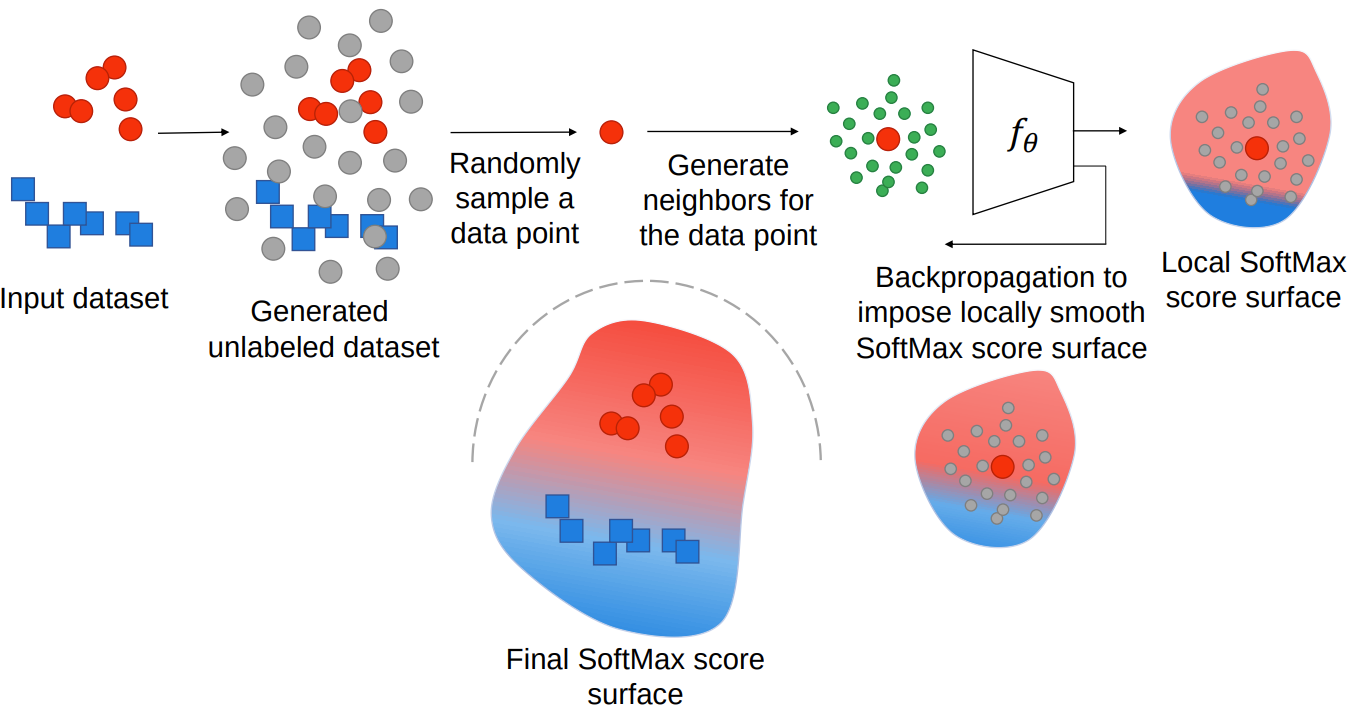
Our code is available at: https://github.com/MahsaPaknezhad/AdversariallyRobustTraining
Please cite our paper:
Paknezhad, M., Ngo, C. P., Winarto, A. A., Cheong, A., Yang, B. C., Jiayang, W., & Kuan, L. H. (2021). Explaining Adversarial Vulnerability with a Data Sparsity Hypothesis. arXiv preprint arXiv:2103.00778.
Whole Slide Image Registration
Whole slide images (WSI) are extremely high resolution microscopic images of multiple 2D slices cut from a tissue volume. The cutting and imaging process deforms the tissue in many ways. Such deformations include tearing, stretching, folding and compressing. Reconstructing the tissue volume by registering (aligning) the whole tissue in the acquired 2D WSI is therefore very complex and often results in poor outcomes. We propose a multi-resolution algorithm that registeres and reconstructs a region of interest in the whole slide images. The regional and multi-resolution approach results in more accurate reconsturction outcomes better than the state-of-the-art algorithms. A diagram of our multi-resolution approach is shown below. The region of interest (ROI) is show by a box in Level0 (highest resolution) of the WSI. The ROI is mapped to lower resolutions (Level1 and Level2) and registration is performed in these ROIs only.
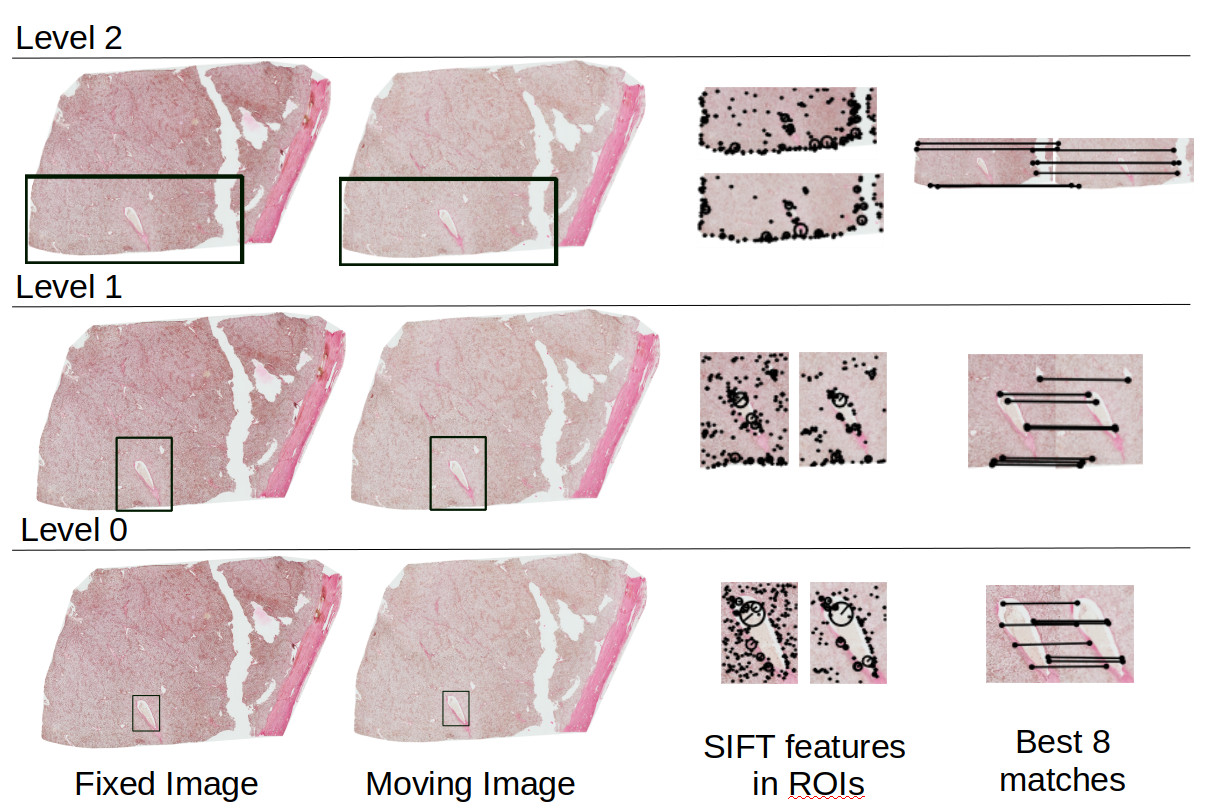
The code is available at the link below:
https://github.com/MahsaPaknezhad/WSIRegistration
Please cite our paper:
Paknezhad, M., Loh, S. Y. M., Choudhury, Y., Koh, V. K. C., Yong, T. T. K., Tan, H. S., … & Lee, H. K. (2020). Regional registration of whole slide image stacks containing major histological artifacts. BMC bioinformatics, 21(1), 1-20.
